Storage structure
This page is documentation of the BCPU storage space on NIRD.
Note
This page is currently under development, so expect frequent changes to the data structure, as it evolves and implementation begins.
Status of restructure
We are currently in the process of creating and migrating to a new directory structure to hold the contents of the NS9039K project space.
Top-level structure
A high-level structure diagram is included below to demonstrate the top-level directories. This includes the directories data, users, projects, repos and www.
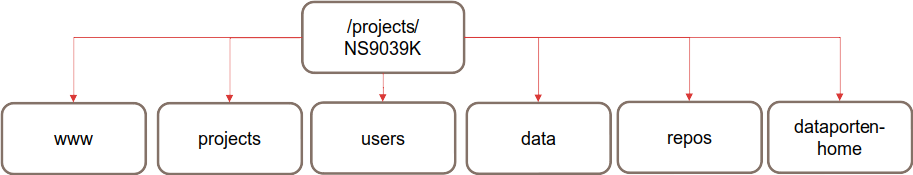
Fig. 1 The top-level data structure.
The purpose and structure of each of these directories is described in their respective sections of this document.
Data directory
The purpose of the data directory is to house data that we use within the BCPU. All datasets should be stored and documented here, following the best practices outlined on this page. This includes derived datasets, which are datasets to which the user has done some amount of processing, but that might still be useful for other users. Note that the data directory has a regulated structure, and so when making changes within this space, the rules and recommendations on this page should be followed.
The data structure is composed of collections and datasets.
A collection is a directory that contains other collections and/or datasets. It should contain a README.json based on the template, and indicate that the directory is a collection by having a value of isCollection:1.
A dataset can contain multiple sub-directories for different variables, time aggregations or ensemble members. It should contain a README.json based on the template, and indicate that the directory is a dataset by having a value of isDataset:1.
The data directory contains two top-level directories; external and internal. The external directory is for model, reanalysis or obervational data which we have downloaded from an external source, for example, the ERA5 reanalysis. The internal directory is for experiments which have been run “in-house”, for example, the output from running a new NorCPM experiment.
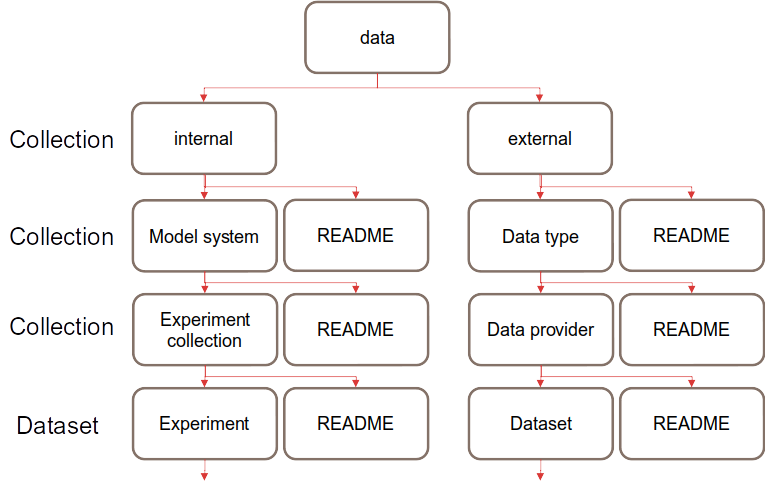
Fig. 2 General structure of the data directory
The general rules that accompany this structure are as follows:
Data must have the same number of directory levels as in Fig. 2. If there is a stand-alone experiment/dataset, it should still be placed in an “Experiment collection” or “Data provider” collection directory even if it is the only experiment/dataset in the collection. There should never be a mixture of collections and datasets in one directory.
As shown in Fig. 2, each collection and dataset directory must contain a README which follows the README template.
Data must be contained in a sub-directory of the dataset level, rather than alongside the README and any other directories. For example, the experiment/dataset may have a large number of ensembles, and these should be organized into one or more sub-directories so as to make the other files in the experiment more discoverable, recommendations for these names are included in the following two sections.
Internal data
We have designed some rules and some recommendations for organizing data from our internal experiments. The general structure of the internal directory can be seen in Fig. 2.
In this structure diagram, there are several required directory layers; Model system, Experiment collection and Experiment, these are all defined as follows:

Model system: the version of the Earth System Model used to carry out experiments. Different model systems are defined by having different reference (historical or piControl) runs.

Experiment collection: a group of experiments which have some similarity, usually if a user is running a set of experiments, they would create their own experiment collection to hold all these experiments together.

Experiment: a single dataset with, for example, a particular forcing or data assimilation scheme.
Fig. 3 shows an example for the internal data structure for one model system. We have provided some recommended directory names in grey and some example sub-directories in gold to complement the general structure. These should be used where possible and appropriate.
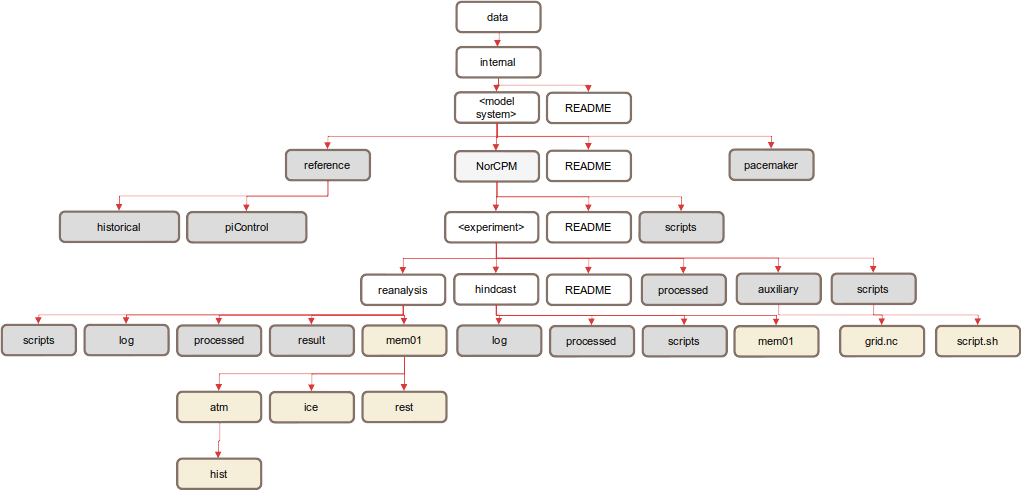
Fig. 3 Example structure of an internal dataset
Some notes on this example:
Where there are NorCPM runs, place them into an ‘experiment collection’ named NorCPM.
For NorCPM experiments, create separate directories hindcast and reanalysis.
Use a reference directory to store reference experiments for a model system, including historical and piControl runs.
Internal dataset sub-directories:
External data
The general structure of the external directory is similar to that of the internal one, it is divided into directories for Data type, Data provider and Dataset as seen in Fig. 2. These terms are defined as follows:

Data type: a collection based on the nature of the data, in practice there are three directories; observation, reanalysis and model.

Data provider: a collection of datasets that are grouped based on data provider. This could be a centre (e.g. ECMWF or NOAA), or a multi-centre project (e.g. CMIP).

Dataset: this is an individual experiment (e.g. rcp45), reanalysis product (e.g. ERA5), or observational records.
External dataset sub-directories:
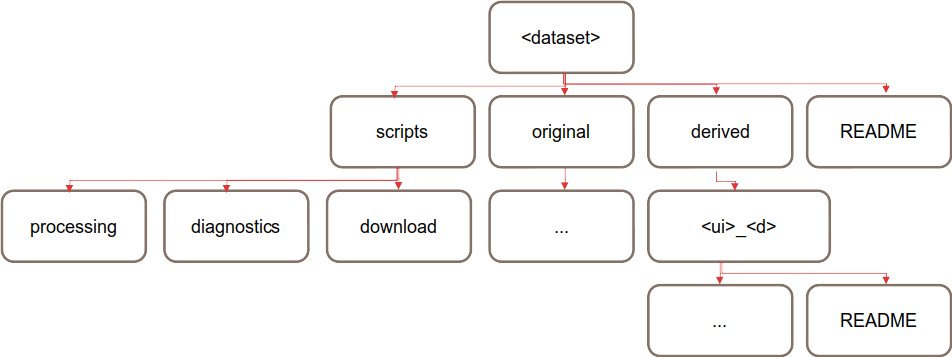
Fig. 4 The structure of an external dataset
original: this is where the original data downloaded from the external source should be kept. This data does not have to be completely unmodified, for example, it could be compressed, or the file format could be converted.
derived/<ui>_<d>: this is where datasets that have undergone some significant changes (e.g. regridding) should be stored. It is recommended to name sub-directories with <ui> as user initials (e.g. tb) and <d> as a brief description (e.g. 1x1).
scripts: code used for downloading the data, preprocessing the data, or even diagnostic scripts can exist in this directory of the data structure.
Users directory
personal code, plots and other files belonging to a user should be placed in a directory within users/. It is recommended that the name of a user’s directory matches their username. The structure of individual user directories is not regulated, and this space can be organized to the user’s convenience.
Projects directory
projects can contain data, scripts and outputs, and it may be convenient to store these related files in one directory, in this case a directory named after the project can be placed in the projects/ directory. The organization of these spaces is the responsibility of the project manager.
Repos directory
local copies of shared Git repositories will reside here, there is a version of all of these files within the BjerknesCPU GitHub.
www directory
this directory contains files to be accessible through the web interface at http://ns9039k.web.sigma2.no/.
More information
For further information, or if you are having issues with the new data structure, please contact BCPU-support, or make an issue in the BjerknesCPU/BCPU-support GitHub repository.